I'm trying to reduce some new data from ALMA's longest baselines (C10, B6).
There are several exciting errors:
- The lines were all flagged out. Thankfully, there is a calibrated_final.ms.backup file that does not have the lines flagged out. flagmanager was unable to restore the flags. The flagging comes from the manual flagging done by the QA2 reducer, which was used to make the continuum image. That's fine, but the unflagging step didn't work. The uvcontsub data, as a result, contain no line data. ?????
- I can't image the full cube because of errors like this:
2021-12-08 08:07:34 INFO SynthesisImagerVi2::defineImage Shape : [9600, 9600, 1, 1920]Spectral : [2.31563e+11] at [0] with increment [976510]
2021-12-08 08:07:34 INFO SynthesisImagerVi2::defineImage Set Gridding options for [S255IR-SMA1_sci.spw1.cube.I.manual] with ftmachine : gridft
2021-12-08 08:07:34 INFO SynthesisImagerVi2::nSubCubeFitInMemory Required memory: 5.468e+05 GB. Available mem.: 166.3 GB (rc, mem. fraction: 70%, memory: -) => Subcubes: 1920. Processes on node: 64.
2021-12-08 08:07:34 INFO SynthesisImagerVi2::weight() Set imaging weights : Briggs weighting: sidelobes will be suppressed over full image
2021-12-08 08:07:34 INFO SynthesisImagerVi2::weight() Doing spectral cube Briggs weighting formula -- norm
2021-12-08 08:07:34 INFO task_tclean::SynthesisDeconvolver::setupDeconvolution Set Deconvolution Options for [S255IR-SMA1_sci.spw1.cube.I.manual] : hogbom
2021-12-08 08:07:34 WARN tclean::::casa Memory available 187904819 kB is very close to amount of required memory 3982754512 kB
2021-12-08 08:07:34 INFO task_tclean::SynthesisImager::makePSF ----------------------------------------------------------- Make PSF ---------------------------------------------
2021-12-08 08:07:34 INFO task_tclean::SynthesisImagerVi2::nSubCubeFitInMemory Required memory: 5.468e+05 GB. Available mem.: 166.3 GB (rc, mem. fraction: 70%, memory: -) => Subcubes: 1920. Processes on node: 64.
Well, there's no error there, but the PSF maker can't handle making even a single frame. It had no trouble making a 9600x9600 MTMFS image for the continuum, though, so it's pretty unclear to me why the cube is getting OOM-killed. My best guess is that the 64 processes are too much for CASA to handle, and I need to run with fewer to "trick" it into not using up that much memory at once. Maybe I just need to make the PSFs in serial mode. I don't want to wait around to make the cubes, let alone clean them, in serial mode.
- Parallel failed with this:
2021-12-08 14:26:19 INFO tclean::::casa ##########################################
2021-12-08 14:26:19 INFO tclean::::casa ##### Begin Task: tclean #####
2021-12-08 14:26:19 INFO tclean::::casa tclean( vis='calibrated_final.ms', selectdata=True, field='S255IR-SMA1', spw='1', timerange='', uvrange='', antenna='', scan='', observation='', intent='', datacolumn='corrected', imagename='S255IR-SMA1_sci.spw1.cube.I.zoom.manual', imsize=[500, 500], cell='0.0042arcsec', phasecenter='', stokes='I', projection='SIN', startmodel='', specmode='cube', reffreq='', nchan=-1, start='', width='', outframe='lsrk', veltype='radio', restfreq=[], interpolation='linear', perchanweightdensity=True, gridder='standard', facets=1, psfphasecenter='', wprojplanes=1, vptable='', mosweight=True, aterm=True, psterm=False, wbawp=True, conjbeams=False, cfcache='', usepointing=False, computepastep=360.0, rotatepastep=360.0, pointingoffsetsigdev=[], pblimit=0.2, normtype='flatnoise', deconvolver='hogbom', scales=[], nterms=2, smallscalebias=0.0, restoration=True, restoringbeam=[], pbcor=True, outlierfile='', weighting='briggs', robust=0.0, noise='1.0Jy', npixels=0, uvtaper=[], niter=10000, gain=0.1, threshold='10mJy', nsigma=0.0, cycleniter=-1, cyclefactor=1.0, minpsffraction=0.05, maxpsffraction=0.8, interactive=False, usemask='user', mask='', pbmask=0.0, sidelobethreshold=3.0, noisethreshold=5.0, lownoisethreshold=1.5, negativethreshold=0.0, smoothfactor=1.0, minbeamfrac=0.3, cutthreshold=0.01, growiterations=75, dogrowprune=True, minpercentchange=-1.0, verbose=False, fastnoise=True, restart=True, savemodel='none', calcres=True, calcpsf=True, psfcutoff=0.35, parallel=True )
2021-12-08 14:26:19 INFO tclean::::casa Verifying Input Parameters
2021-12-08 14:26:19 INFO SynthesisImagerVi2::selectData MS : calibrated_final.ms | Selecting on fields : S255IR-SMA1 | Selecting on spw :1 | [Opened in readonly mode]
2021-12-08 14:26:19 INFO SynthesisImagerVi2::selectData NRows selected : 0
2021-12-08 14:26:19 SEVERE tclean::::casa Task tclean raised an exception of class RuntimeError with the following message: Parallel transport layer not initialized
2021-12-08 14:26:19 INFO tclean::::casa Task tclean complete. Start time: 2021-12-08 09:26:19.103483 End time: 2021-12-08 09:26:19.314353
2021-12-08 14:26:19 INFO tclean::::casa ##### End Task: tclean #####
2021-12-08 14:26:19 INFO tclean::::casa ##########################################
I'm guessing this is a bad error message and the real error is that the data selection failed. Probably when I copied over the continuum_final.ms.backup file, it uses the original numbering (spw 25, 27, etc.) instead of the new numbering. I guess the renumbering is cased by uvcontsub, which I'm now skipping because it may have broken my data and because I'd rather image the continuum. That proved correct; the error message above just means that I selected a nonexistent SPW.
Flagged Out Lines
After re-copying over the calibrated MS and confirming that no lines were flagged out, I re-imaged... and the lines are again missing.
# show the flags graphically
flagdata(vis='calibrated_final.ms', mode='summary', spwchan=True, display='both')
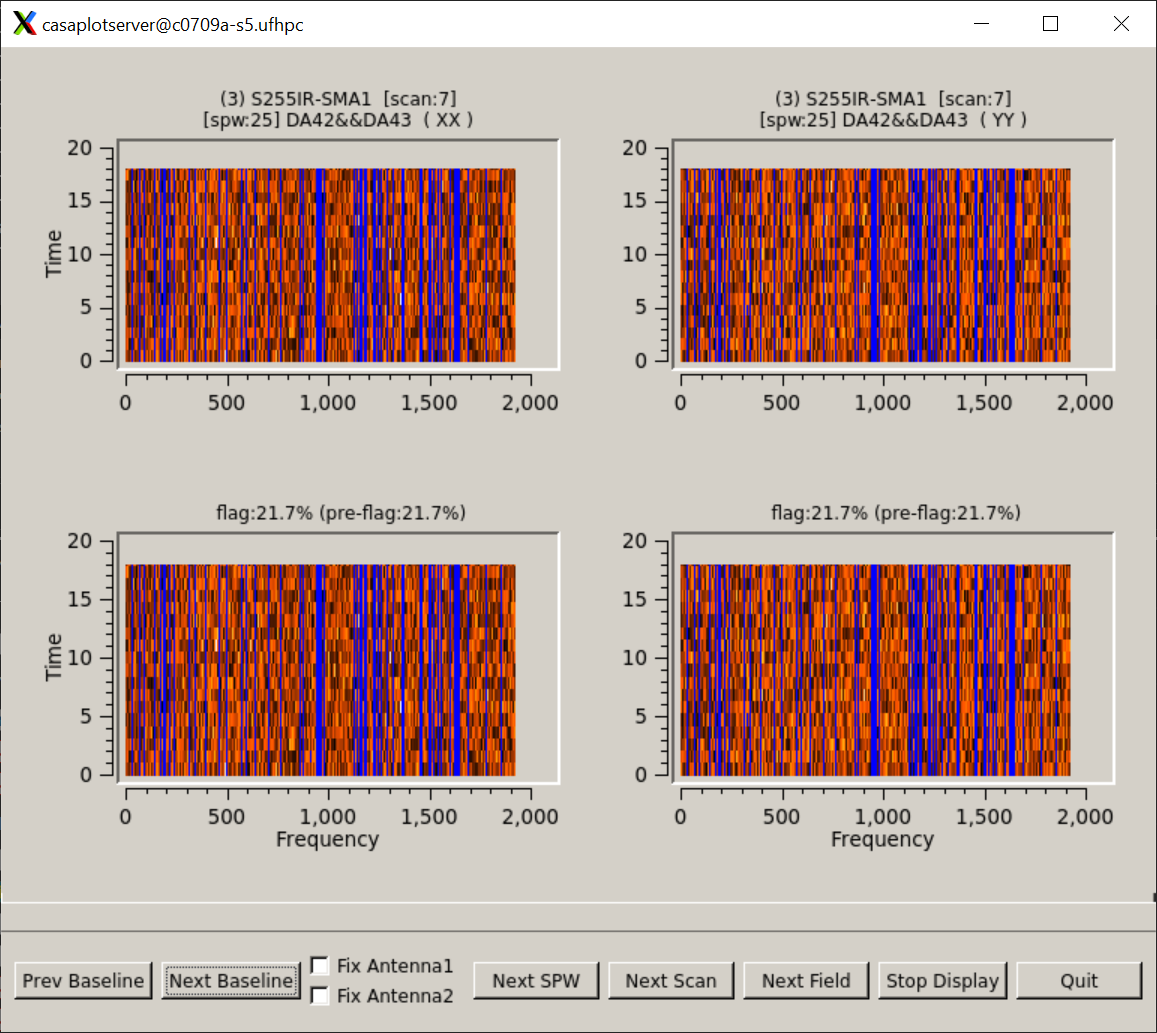
The flags are coming from tclean, which makes no sense.
There are messages like these:
2021-12-08 15:27:19 WARN MPICommandServer::command_request_handler_service::SIImageStore::getPSFGaussian::MPIServer-51 (file src/code/synthesis/ImagerObjects/SIImageStore.cc, line 2037) PSF is blank for[C9:P0] [C10:P0] [C11:P0] [C12:P0] [C13:P0] [C14:P0] [C15:P0] [C16:P0] [C17:P0] [C23:P0] [C24:P0] [C25:P0] [C26:P0]
scattered throughout the PSF making.
The PSF spectrum ends up like this:
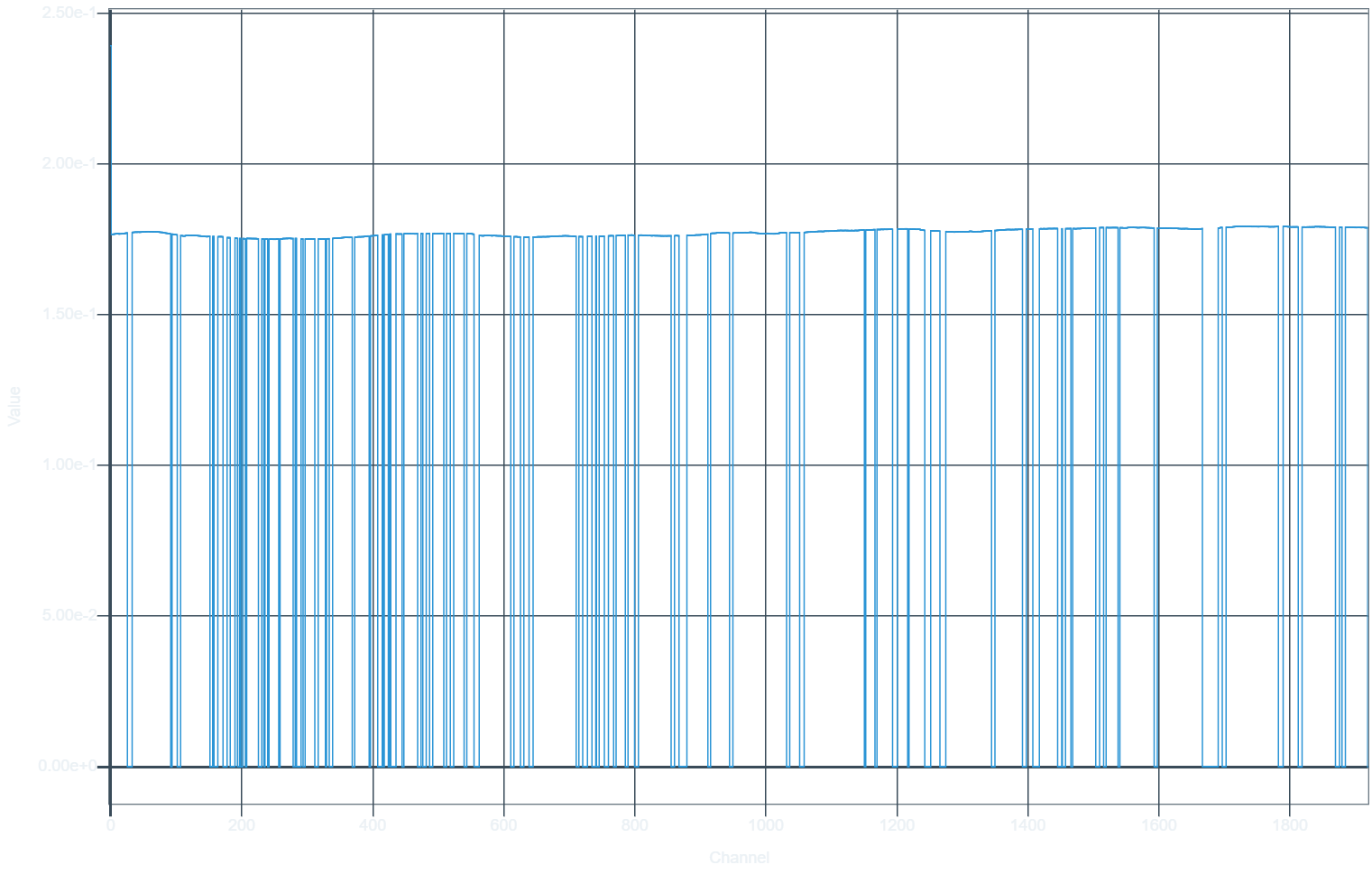
Channels are totally flagged out. But in the data, they are not:
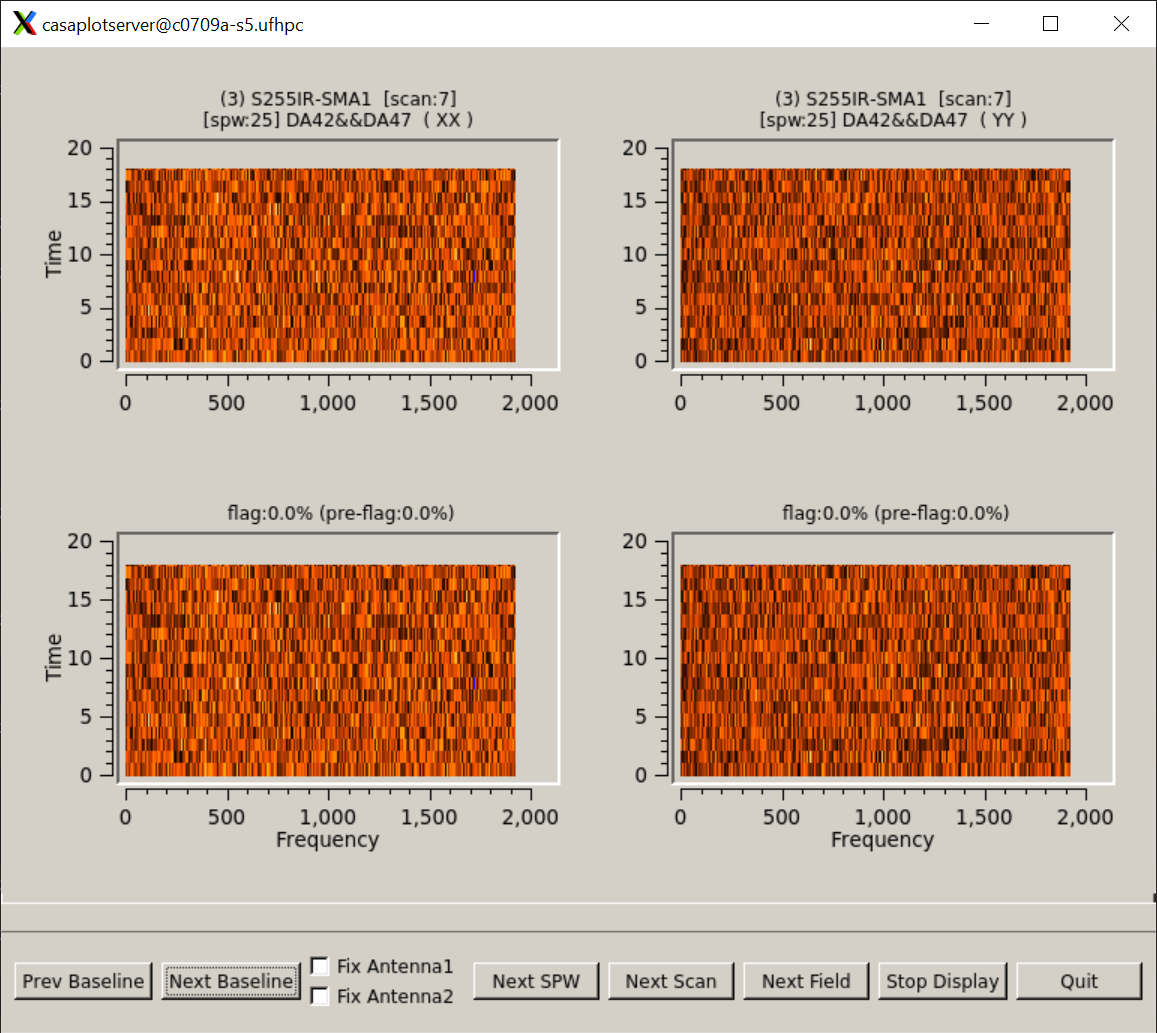
Running the tclean without parallel, i.e. with parallel=False, resulted in a smooth, unbroken PSF.
So, if we dig back into what happened in the parallel tclean....
2021-12-08 15:26:12 INFO SynthesisImagerVi2::selectData NRows selected : 848700
2021-12-08 15:26:15 INFO SynthesisImagerVi2::defineImage Define image coordinates for [S255IR-SMA1_sci.spw0.cube.I.zoom.manual] :
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs phaseCenter='Direction: [-0.0535075, 0.949606, 0.308847]' Channels equidistant in freq
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs+ Central frequency (in output frame) = 2.345e+11 Hz
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs+ Width of central channel (in output frame) = 976510 Hz
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs+ Number of channels = 1920
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs+ Total width of SPW (in output frame) = 1.8749e+09 Hz
2021-12-08 15:26:16 INFO MSTransformRegridder::calcChanFreqs+ Lower edge = 2.33563e+11 Hz, upper edge = 2.35437e+11 Hz
2021-12-08 15:26:16 INFO SynthesisImagerVi2::defineImage Impars : start
2021-12-08 15:26:16 INFO SynthesisImagerVi2::defineImage Shape : [500, 500, 1, 1920]Spectral : [2.33563e+11] at [0] with increment [976510]
2021-12-08 15:26:16 INFO SynthesisImagerVi2::defineImage Set Gridding options for [S255IR-SMA1_sci.spw0.cube.I.zoom.manual] with ftmachine : gridft
2021-12-08 15:26:16 INFO SynthesisImagerVi2::nSubCubeFitInMemory Required memory: 1483 GB. Available mem.: 173.6 GB (rc, mem. fraction: 70%, memory: -) => Subcubes: 9. Processes on node: 64.
2021-12-08 15:26:16 INFO SynthesisImagerVi2::weight() Set imaging weights : Briggs weighting: sidelobes will be suppressed over full image
2021-12-08 15:26:16 INFO SynthesisImagerVi2::weight() Doing spectral cube Briggs weighting formula -- norm
2021-12-08 15:26:16 INFO task_tclean::SynthesisDeconvolver::setupDeconvolution Set Deconvolution Options for [S255IR-SMA1_sci.spw0.cube.I.zoom.manual] : hogbom
2021-12-08 15:26:17 INFO task_tclean::SynthesisImager::makePSF ----------------------------------------------------------- Make PSF ---------------------------------------------
... a bunch of stuff ...
2021-12-08 15:27:11 INFO MPICommandServer::command_request_handler_service::SynthesisImagerVi2::makePrimaryBeam::MPIServer-54 vi2 : Evaluating Primary Beam model onto image grid(s)
2021-12-08 15:27:12 INFO MPICommandServer::command_request_handler_service::SynthesisImagerVi2::makePrimaryBeam::MPIServer-38 vi2 : Evaluating Primary Beam model onto image grid(s)
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::calcSensitivity::MPIServer-54 [S255IR-SMA1_sci.spw0.cube.I.zoom.manual] Theoretical sensitivity (Jy/bm):c0:none c1:none c2:none c3:none c4:none c5:none c6:none c7:none c8:none c9:none c10:none c11:none c12:none c13:0.00121258 c14:0.00121257 c15:0.00121255 c16:none c17:none c18:none c19:none c20:none c21:none c22:none c23:none c24:none c25:none c26:0.00121253 c27:0.00121253 c28:0.00121252 c29:0.00121251
2021-12-08 15:27:13 WARN MPICommandServer::command_request_handler_service::SIImageStore::getPSFGaussian::MPIServer-54 (file src/code/synthesis/ImagerObjects/SIImageStore.cc, line 2037) PSF is blank for[C0:P0] [C1:P0] [C2:P0] [C3:P0] [C4:P0] [C5:P0] [C6:P0] [C7:P0] [C8:P0] [C9:P0] [C10:P0] [C11:P0] [C12:P0] [C16:P0] [C17:P0] [C18:P0] [C19:P0] [C20:P0] [C21:P0] [C22:P0] [C23:P0] [C24:P0] [C25:P0]
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::getPSFGaussian::MPIServer-54 Time to fit Gaussian to PSF 0.12
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::printBeamSet::MPIServer-54 Restoring Beams
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::printBeamSet::MPIServer-54 + Pol Type Chan Freq Vel
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::printBeamSet::MPIServer-54 + I Max 0 2.351450e+11 -824.59 0.0254 arcsec x 0.0197 arcsec pa= -6.8506 deg
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::printBeamSet::MPIServer-54 + I Min 1 2.351460e+11 -825.84 0.0000 arcsec x 0.0000 arcsec pa= 0.0000 deg
2021-12-08 15:27:13 INFO MPICommandServer::command_request_handler_service::SIImageStore::printBeamSet::MPIServer-54 + I Median 9 2.351538e+11 -835.82 0.0254 arcsec x 0.0197 arcsec pa= -6.8506 deg
These blank psfs are wrong, there are no flagged data in this data set (no flagged channels). Something about the parallel version of clean is reading the data wrong.
Solution
I finally solved this by manually unflagging the channels. Thanks to Brian Svoboda for helping with the antenna selection syntax.
flagdata(linevis, spw=str(orig_spw), antenna='*&*', mode='unflag')
I don't understand why flagdata was showing the channels as being unflagged, as they were clearly flagged in all data sets (original & backup). I don't understand _when_ they were flagged, either, as it seems they were flagged even before continuum imaging was done.